Research

The research in our lab aims to understand the mechanisms by which non-coding RNAs regulate gene expression. To address these questions, our lab has developed numerous tools to study lncRNA biology – including biochemical methods to comprehensively define proteins that interact with a specific RNA, molecular methods to map RNAs to chromatin and pre-mRNA, and genomic methods for mapping higher-order 3D organization of RNA and DNA in the nucleus.
Our work has uncovered the detailed molecular mechanisms that enable Xist to exploit 3-dimensional proximity to identify its target sites on the X chromosome, interact with the SHARP/SMRT/HDAC3 complex to exclude RNA Polymerase II and silence transcription, and remodel the 3-dimensional structure of the X chromosome to enable chromosome-wide RNA spreading and silencing. These results have led to new paradigms in ncRNA biology, 3D genome organization, and gene regulation.
It has long been thought that nuclear-retained RNAs might play a role in organizing the structure of the nucleus. Yet, the identity and possible mechanism for this RNA component of nuclear organization remains a mystery.
Over the last few years it has become clear that mammalian genomes encode many thousands of long ncRNAs (lncRNAs) that are generally enriched within the chromatin-associated fraction of the nucleus, are expressed in a highly cell-type specific manner, and play important roles in gene regulation.
We have recently shown that hundreds of non-coding RNAs form stable, high concentration, spatially localized compartments throughout the nucleus that bind and recruit diffusible protein molecules. Importantly, many RNA binding proteins (RBPs) contain intrinsically disordered regions (IDRs) which facilitate self-aggregation, and in several cases, RNA has been shown to facilitate this phase separation process. Thus, ncRNAs may act to facilitate phase transitions by increasing spatial concentrations of such RBPs, or by directly participating in heterotypic multivalent interactions through RNA sequence repeats. In this way, specific ncRNAs provide a valuable platform for dissecting the function and mechanism of nuclear organization in regulating gene expression.
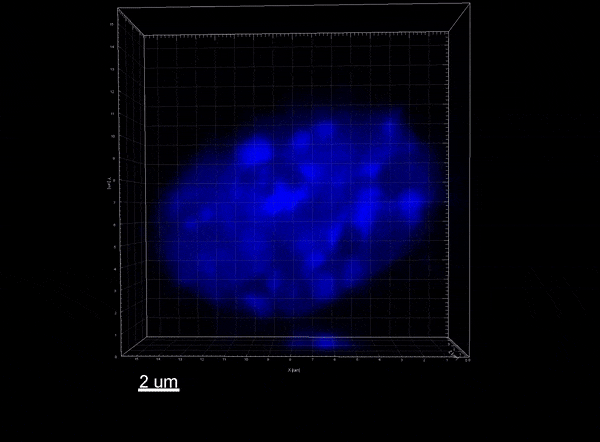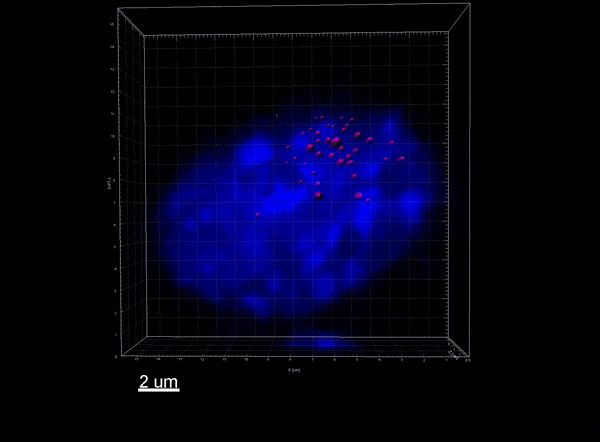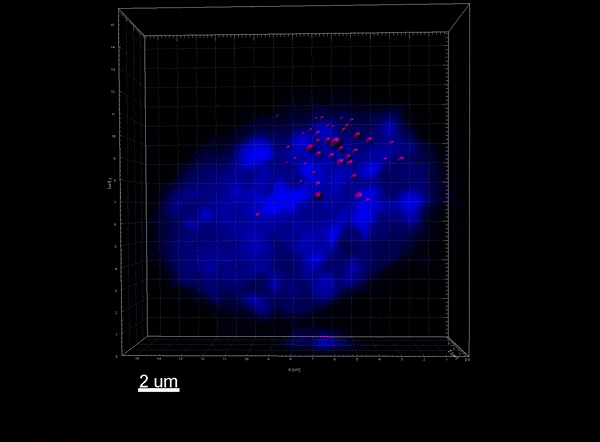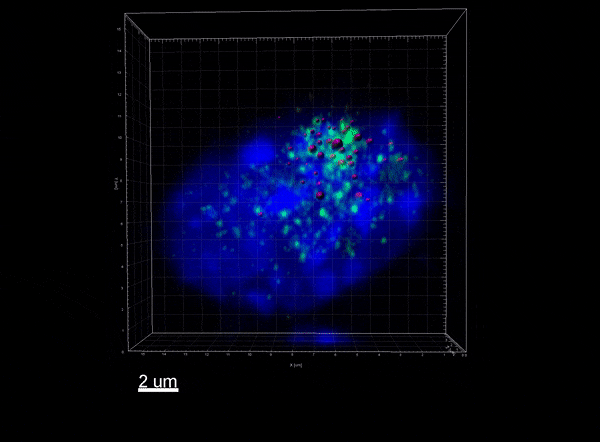
The Xist lncRNA (magenta) recruits the SHARP protein (green) to silence the X chromosome.

Active Research
Our lab is developing novel methods based on our published SPRITE approach to enable comprehensive mapping of RNA, DNA, and protein assemblies in the nucleus in a manner that generates direct information about 3-dimensional interactions and provides single molecule resolution of complexes along simultaneous information about transcription. We are using these methods along with genetic perturbation, biochemistry, and microscopy to dissect the role of ncRNA-mediated processes that link dynamic 3D nuclear structure and gene regulation in biology and disease.
Guttman Lab is investigating key mechanisms and unique roles that lncRNAs can play in gene regulation.
Dynamic assembly and disassembly of functional compartments.
Spatial partitioning of the nucleus for efficient regulation.
Spatial amplification of regulatory information.
Phase transitions and maintenance of regulatory states.
We are studying these properties within several key biological contexts, including:
- Chromosome-wide epigenetic silencing,
- Kinetic coupling of mRNA transcription and splicing
- RNA-induced aggregation and cellular toxicity in neurodegenerative disorders.
We expect that the insights derived will be broadly applicable to many other regulatory processes and will provide an important framework for understanding the roles that biochemical compartmentalization plays in the quantitative control of gene regulation.

Previous Research
Xist Silencing and Chromatin Organization
We uncovered the molecular mechanisms by which the Xist ncRNA leads to chromosome-wide transcriptional silencing. Specifically, we demonstrated that Xist exploits 3-dimensional proximity to identify target sites on the X chromosome, remodels the 3-dimensional structure of the X chromosome to spread across it, and interacts with the SHARP/SMRT/HDAC3 complex to exclude RNA Polymerase II and silence transcription at each gene.
We have shown that the mechanism by which Xist seeds the formation of a nuclear compartment on the X chromosome represents a central and widespread role for ncRNAs in shaping nuclear structure. Specifically, we found that hundreds of ncRNAs form stable compartments in the nucleus by accumulating at a high concentration in proximity to their transcription loci (seed), binding to specific diffusible proteins (bind), and recruiting these proteins to form regulatory compartments (recruit). By increasing spatial concentrations of regulatory proteins, ncRNAs can facilitate concentration-dependent phase transitions to amplify the concentration and function of specific regulatory proteins in the nucleus.
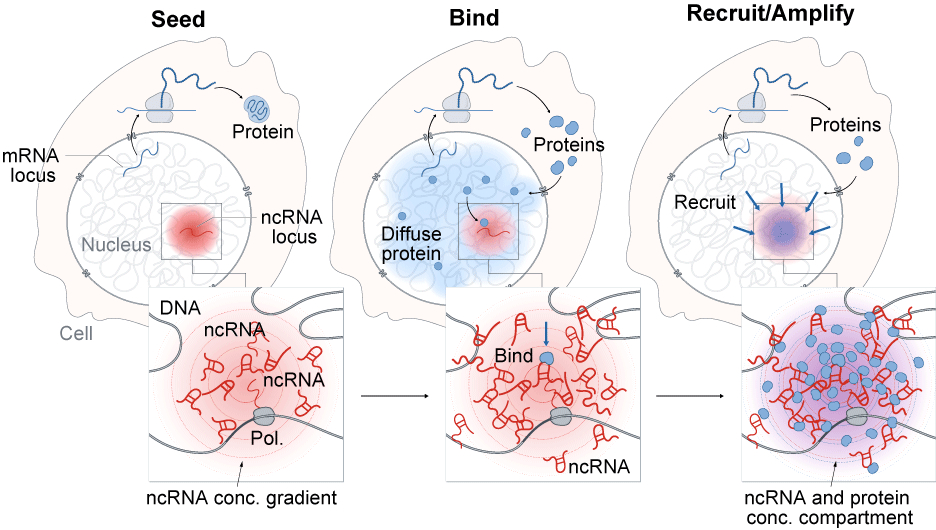
A general model by which ncRNAs drive the formation of nuclear compartments. The process of ncRNA transcription creates a concentration gradient of ncRNA transcript with the highest concentrations near its transcriptional locus (SEED, left). Because these RNAs are functional immediately upon transcription and can bind with high affinity to diffusible RNAs and proteins (BIND, middle panel), they can act to change the dynamic equilibrium of these proteins to concentrate them in a spatial compartment (RECRUIT, right panel).
Nuclear Compartmentalization
We have shown that ncRNA-mediated nuclear compartments play essential roles in a wide range of essential nuclear functions, including RNA processing, heterochromatin organization, and gene regulation. Our results have established that nuclear ncRNAs are uniquely suited to shape nuclear structure and have established new paradigms in lncRNA biology, 3D genome organization, and gene regulation. We are actively exploring these roles more generally to understand how they may explain critical aspects of gene regulation.
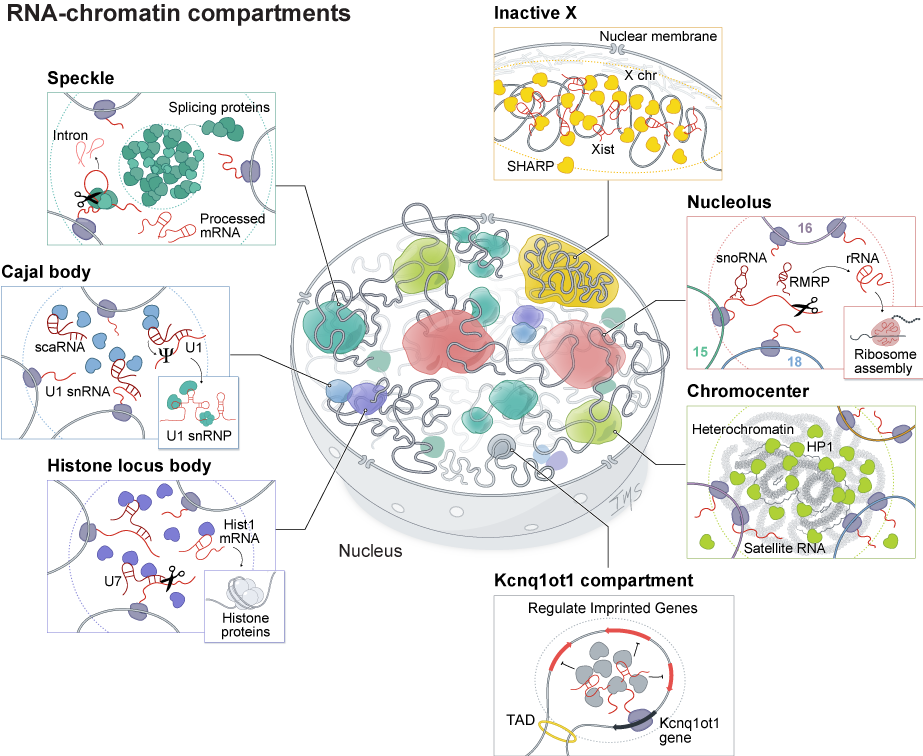
RNA-mediated compartments regulate various processes throughout the nucleus. RNA has been shown to organize nuclear bodies involved in RNA processing, chromatin regulation, and gene expression. These include RNA processing bodies such as: Nucleolus: The site of ribosome biogenesis within the nucleus. Histone locus bodies: HLBs contain histone gene loci and are sites of histone pre-mRNA transcription and processing. Speckles: Nuclear speckles are nuclear bodies with high concentrations of pre-mRNA splicing proteins and snRNAs. Inactive X chromosome: The Xist long non-coding RNA (lncRNA) is responsible for chromosome-wide silencing, compaction, and localization of the inactive X chromosome to the nuclear lamina in female cells. Chromocenter: Satellite DNA repeats on multiple chromosomes are compacted into heterochromatin-dense foci. Genomic imprinting: The Kcnq1ot1 lncRNA silences gene expression of imprinted target genes located in a 3D compartment next to its transcriptional locus.
SARS-CoV-2 Viral Proteins
In response to the Covid19 pandemic, we have applied our expertise and methods for studying RNA-protein interactions to understand the molecular basis of SARS-CoV-2 infection and pathogenesis. We comprehensively defined the interactions between each SARS-CoV-2 protein and human RNAs. We showed that viral proteins disrupt three essential steps of protein production – mRNA splicing, protein translation, and membrane and secretory protein trafficking – and in this way suppress the innate immune response to viral infection.
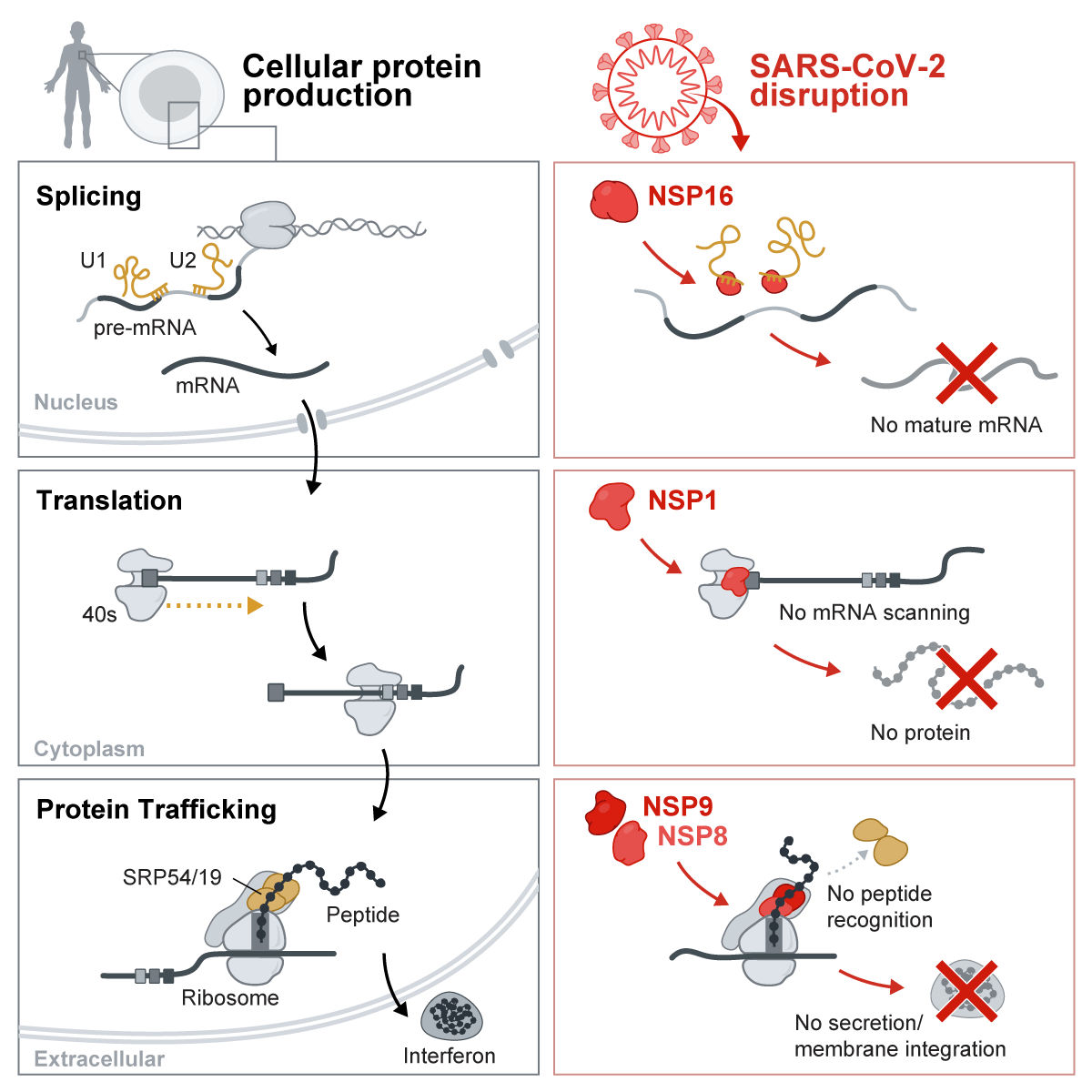
SARS-CoV-2 utilizes a multipronged strategy to antagonize essential cellular processes to suppress host defenses. These include binding of NSP16 binds to the mRNA binding domains of the U1 and U2 splicing RNAs which acts to suppress global mRNA splicing. NSP1 binds to 18S ribosomal RNA in the mRNA entry channel of the ribosome and leads to global inhibition of mRNA translation upon infection. Finally, NSP8 and NSP9 bind to the 7SL RNA in the Signal Recognition Particle and interfere with protein trafficking to the cell membrane upon infection. Disruption of each of these essential cellular functions acts to suppress the interferon response to viral infection.

