Guttman Lab
Decoding the roles of non-coding RNAs in shaping nuclear structure and gene expression.
We are an integrated team of experimental and computational biologists who work together to understand how nuclear compartments control gene expression programs and cell state decisions. We believe that the strength of our science arises from the diversity of our ideas and experiences. We are committed to fostering an inclusive and diverse laboratory culture and as such welcome students, postdocs, and visiting scholars from all backgrounds to join our group.

Our Lab
We seek to understand fundamental principles of biological regulation within the nucleus. Our team of scientists from diverse backgrounds allows us to approach these questions from a holistic perspective with an integrated strategy. We do not make a clear distinction between experimental and computational projects; our projects are goal-centered and use all tools critical for success. When necessary, we develop new tools to answer questions that were previously inaccessible and apply these tools to decipher new paradigms and improve our understanding of essential biological functions.
In The News
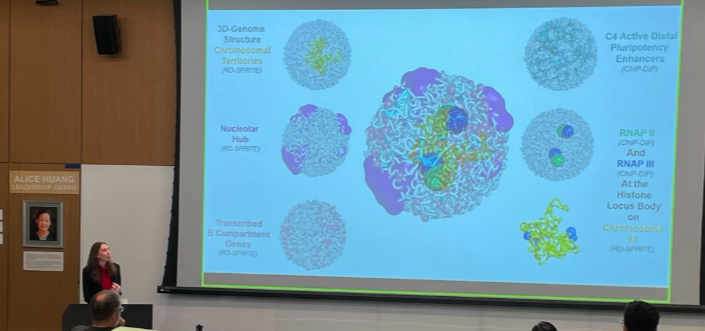
Revealing chromosome contours, one dot at a time
Nature interviews Guttman Lab in this week’s Technology Feature discussing the latest technological advances for studying 3D genome organization. In concert with the in situ microscopy renderings created at Caltech’s Cai Lab, Guttman Lab’s SPRITE method has opened the door to discovery driven proximity interactions – even long distance interactions. Even lab mascot SHARP-y made the feature! Read the full article at Nature.com
The Vast Little Library Inside of Your Cells
Featured this week in Caltech News: Using the latest innovation developed at Guttman Lab, RNA-DNA SPRITE, our team discovered that non-coding RNA create “compartments” within the nucleus and shepherd key molecules to precise regions in the genome. Read this review now at Caltech News
Prashant awarded de Karman Fellowship
Reserved for graduate students going into their final year of doctoral work, the fellowship was created to recognize and support outstanding scholastic achievement. Read about Prashant’s work and the award at Caltech News
Announcements
Farewell to Our Outstanding Summer Students!
Farewell to Our Outstanding Summer Students! August 2023 was a bittersweet moment as we bid farewell to our talented summer students, Jessica Mann (Amgen Scholars, MIT ’24) and Tanvi Ganapathy (SURF, Caltech ’26).
Baking Up Fun at Guttman Lab’s Summer Cookie Hour Challenges!
Baking Up Fun at Guttman Lab’s Summer Cookie Hour Challenges! Every Tuesday in the Guttman lab we host our summer series of weekly cookie hour baking challenges. Our lab members come together for friendly bake-offs, treats, tea, and the crowning of the Guttman Lab Bakeoff winner. It’s a delightful way to foster team spirit and satisfy our sweet tooth!
Isabel Goronzy Triumphs in Thesis Defense!
Summer lab retreat at Descanso Gardens
Guttman Lab Shines at Graduation Day!
Celebrating Prashant Bhat’s Thesis Defense and Everhart Distinguished Student Lecture!
Celebrating Prashant Bhat’s Thesis Defense and Everhart Distinguished Student Lecture! On May 3rd, Prashant Bhat successfully defended his thesis and delivered the prestigious Everhart Distinguished Student Lecture. We’re proud of his achievements and look forward to his year as a postdoctoral scholar in our lab before he heads to medical school at UCLA in July 2024.