Publications
2024
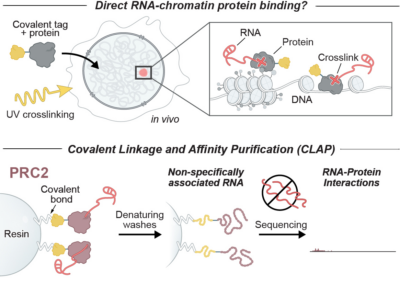
Denaturing purifications demonstrate that PRC2 and other widely reported chromatin proteins do not appear to bind directly to RNA in vivo
Molecular Cell
Jimmy K. Guo*, Mario R. Blanco*, Ward G. Walkup IV, Grant Bonesteele, Carl R. Urbinati, Abhik K. Banerjee, Amy Chow, Olivia Ettlin, Mackenzie Strehle, Parham Peyda, Enrique Amaya, Vickie Trinh, and Mitchell Guttman
2023
ChIP-DIP: A multiplexed method for mapping hundreds of proteins to DNA uncovers diverse regulatory elements controlling gene expression
bioRxiv (preprint)
, , , ,
SPIDR: a highly multiplexed method for mapping RNA-protein interactions uncovers a potential mechanism for selective translational suppression upon cellular stress
bioRxiv (preprint)
Erica Wolin*, Jimmy K. Guo*, Mario R. Blanco, Andrew A. Perez, Isabel N. Goronzy, Ahmed A. Abdou, Darvesh Gorhe, Mitchell Guttman, Marko Jovanovic
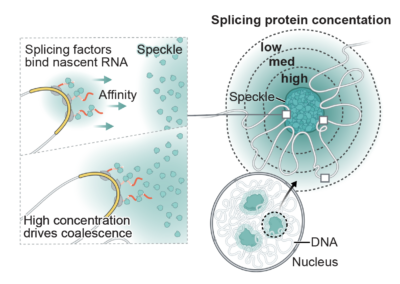
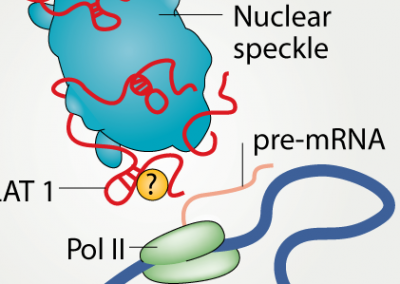
3D genome organization around nuclear speckles drives mRNA splicing efficiency
BioRxiv (preprint)
Prashant Bhat, Amy Chow, Benjamin Emert, Olivia Ettlin, Sofia A. Quinodoz, Yodai Takei, Wesley Huang, Mario R. Blanco, Mitchell Guttman
2022
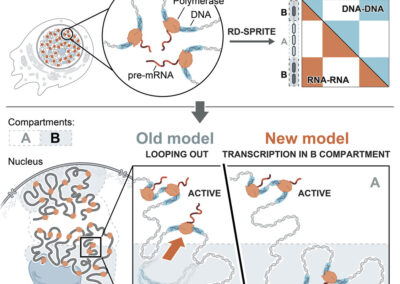
Simultaneous mapping of 3D structure and nascent RNAs argues against nuclear compartments that preclude transcription
Cell Reports
Isabel N. Goronzy*, Sofia A. Quinodoz*, Joanna W. Jachowicz**, Noah Ollikainen**, Prashant Bhat, and Mitchell Guttman
Regulatory non-coding RNAs: everything is possible, but what is important?
Nature Methods
Jimmy K. Guo and Mitchell Guttman
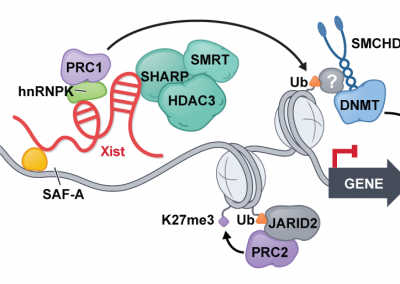
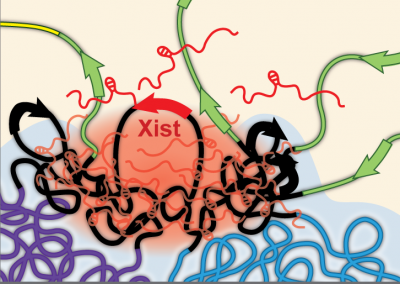
Xist spatially amplifies SHARP recruitment to balance chromosome-wide silencing and specificity to the X chromosome
Nature Structural and Molecular Biology
Joanna W. Jachowicz*, Mackenzie Strehle*, Abhik K. Banerjee, Mario R. Blanco, Jasmine Thai, and Mitchell Guttman
News and Views in: Nature Structural and Molecular Biology
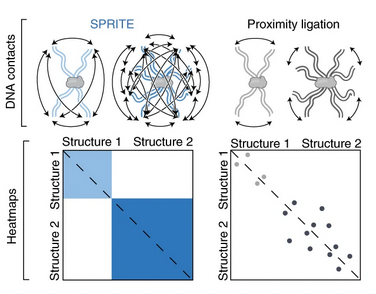
SPRITE: a genome-wide method for mapping higher-order 3D interactions in the nucleus using combinatorial split-and-pool barcoding
Nature Protocols
Sofia A. Quinodoz, Prashant Bhat, Peter Chovanec, Joanna W. Jachowicz, Noah Ollikainen, Elizabeth Detmar, Elizabeth Soehalim, and Mitchell Guttman
2021
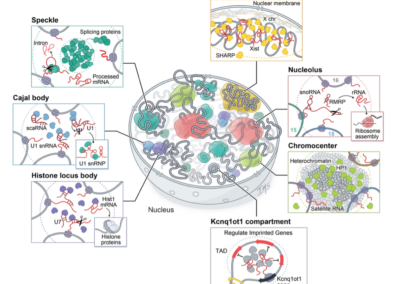
RNA promotes the formation of spatial compartments in the nucleus
Cell
Sofia A. Quinodoz, Joanna W. Jachowicz*, Prashant Bhat*, Noah Ollikainen*, Abhik K. Banerjee**, Isabel N. Goronzy**, Mario R. Blanco, Peter Chovanec, Amy Chow, Yolanda Markaki, Jasmine Thai, Kathrin Plath, and Mitchell Guttman
Featured in: Cell | Nature Reviews Genetics
Single-cell measurement of higher-order 3D genome organization with scSPRITE
Nature Biotechnology
Mary V. Arrastia, Joanna W. Jachowicz, Noah Ollikainen, Matthew S. Curtis, Charlotte Lai, Sofia A. Quinodoz, David A. Selck, Rustem F. Ismagilov & Mitchell Guttman (2021).
Featured in: Nature Methods Research Highlight and Methods to Watch | Epigenie
Essential Roles for RNA in Shaping Nuclear Organization
The Nucleus (2nd edition)
Sofia A. Quinodoz & Mitchell Guttman (2021).
Nuclear compartmentalization as a mechanism of quantitative control of gene expression
Nature Reviews Molecular Cell Biology
Prashant Bhat, Drew Honson, and Mitchell Guttman (2021).
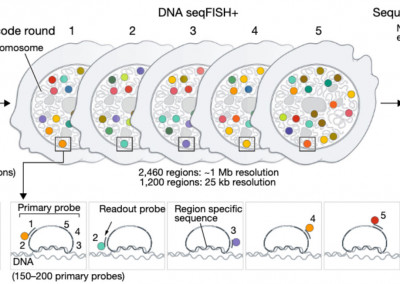
Global architecture of the nucleus in single cells by DNA seqFISH+ and multiplexed immunofluorescence
Nature
Takei Y, Ollikainen N, Zheng S, Pierson N, White J, Shah S, Thoimassie J, Eng L, Guttman M, Yuan GC, Cai, L (2021).
2020
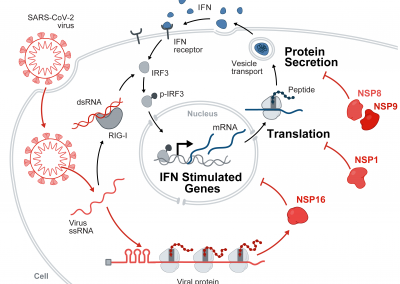
SARS-CoV-2 suppresses mRNA splicing, translation, and protein trafficking in a multipronged mechanism to evade host defenses
Cell
Banerjee AK, Blanco MR, Bruce EA, Honson DD, Chen L, Chow A, Bhat P, Ollikainen N, Quinodoz SA, Loney C, Thai J, Miller ZD, Lin AE, Schmidt MM, Stewart DG, Goldfarb D, De Lorenzo G, Rihn SJ, Voorhees R, Botten JW, Majumdar D, and Guttman M (2020)
High-resolution mapping of multiway enhancer promoter interactions regulating pathogen detection
Molecular Cell
Vangala P, Murphy R, Quinodoz SA, Gellatly K, McDonel PE, Guttman M, Garber M (2020).
An Xist-dependent protein assembly mediates Xist localization and gene silencing
Nature
Pandya-Jones A, Markaki Y, Serizay J, Chitiashvilli T, Mancia W, Damianov A, Chronis C, Papp B, Chen CK, McKee R, Wang XJ, Chau A, Leonhardt H, Zheng S, Guttman M, Black DL, Plath K (2020).
Xist drives spatial compartmentalization of DNA and protein to orchestrate initiation and maintenance of X inactivation
Current Opinions in Cell Biology
Strehle M and Guttman M (2020).
2019
Phase separation drives X-chromosome inactivation: a hypothesis
Nature Structural and Molecular Biology
Cerase A, Armaos A, Neumayer C, Avner P, Guttman M, Tartaglia GG
Approaches for Understanding the Mechanisms of Long Noncoding RNA Regulation of Gene Expression
RNA Worlds: New Tools for Deep Exploration edited by Thomas R. Cech, Joan A. Steitz, and John F. Atkins
Patrick McDonel and Mitchell Guttman (2019)
The bipartite TAD organization of the X-inactivation center ensures opposing developmental regulation of Tsix and Xist
Nature Genetics
van Bemmel JG, Galupa R, Gard C, Servant N, Picard C, Davies J, Szempruch AJ, Zhan Y, Żylicz JJ, Nora EP, Lameiras S, de Wit E, Gentien D, Baulande S, Giorgetti L, Guttman M, Hughes JR, Higgs DR, Gribnau J, Heard E (2019)
2018
Higher-order inter-chromosomal hubs shape 3-dimensional genome organization in the nucleus
Cell
Quinodoz SA, Ollikainen N, Tabak B, Palla A, Schmidt JM, Detmar E, Lai M, Shishkin A, Bhat P, Trinh V, Aznauryan E, Russell P, Cheng C, Jovanovic M, Chow A, McDonel P, Garber M, and Guttman M (2018).
Featured in: Nature Methods | Epigenie
The NORAD lncRNA assembles a topoisomerase complex critical for genome stability
Nature
Munschauer M, Nguyen CT, Sirokman K, Hartigan CR, Hogstrom L, Engreitz JM, Ulirsch JC, Fulco CP, Subramanian V, Chen J, Schenone M, Guttman M, Carr SA, Lander ES (2018).
2017
The 4D nucleome project
Nature
Dekker J, Belmont AS, Guttman M, Leshyk VO, Lis JT, Lomvardas S, Mirny LA, O’Shea CC, Park PJ, Ren B, Politz JCR, Shendure J, Zhong S (2017).
RAP-MS: A Method to Identify Proteins that Interact Directly with a Specific RNA Molecule in Cells
Methods Molecular Biology
McHugh CA and Guttman M (2017).
Re-evaluating the foundations of lncRNA-Polycomb function
EMBO Journal
Blanco MR and Guttman M (2017)
Linking Protein and RNA Function within the same gene
Cell
Szempruch A and Guttman M (2017).
2016
Xist recruits the X chromosome to the nuclear lamina to enable chromosome-wide silencing
Science
Chen CK, Blanco M, Jackson C, Aznauryan E, Ollikainen N, Surka C., Chow A, Cerase A, McDonel PE, Guttman M (2016).
Featured in: Caltech
Long non-coding RNAs (lncRNAs) as spatial amplifiers that control nuclear architecture and gene expression
Nature Reviews Molecular Cell Biology
Engreitz JM, Ollikainen N, Guttman M (2016).
Quantitative predictions of protein interactions with long non-coding RNAs
Nature Methods
Cirillo D, Blanco MR, Armaos A, Buness A, Avner P, Guttman M, Cerase A, Tartaglia GG (2016).
Local regulation of gene expression by lncRNA promoters, transcription, and splicing
Nature
Engreitz JM, Haines JE, Perez EM, Munson G, Chen J, Kane M, McDonel PE, Guttman M, Lander ES (2016).
m6A RNA methylation promotes XIST-mediated transcriptional repression
Nature
Patil DP, Chen CK, Pickering BF, Chow A, Jackson C, Guttman M, Jaffrey SR (2016).
Enhanced CLIP (eCLIP) enables robust and scalable transcriptome-wide discovery and characterization of RNA binding protein binding sites
Nature Methods
Van Nostrand EL, Pratt GA, Shishkin AA, Gelboin-Burkhart C, Fang M, Sundararaman B, Blue SM, Nguyen TB, Surka C, Elkins K, Stanton R, Rigo F, Guttman M, Yeo GW (2016).
Evolutionary analysis across mammals reveals distinct classes of long non-coding RNAs
Genome Biology
Chen J, Shishkin AA, Zhu X, Kadri S, Maza I, Guttman M, Hanna JH, Regev A, Garber M (2016).
2015
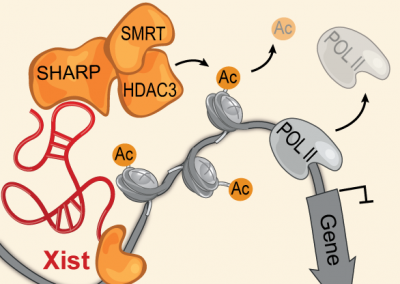
The Xist lncRNA directly interacts with SHARP to silence transcription through HDAC3
Nature
McHugh CA, Chen CK, Chow A, Surka CF, Tran C, McDonel P, Pandya-Jones A, Blanco MR, Burghard C, Moradian A, Sweredoski MJ, Shishkin AA, Su J, Lander ES, Hess S, Plath K, and Guttman M (2015).
Featured in: Nature | Nature Reviews Molecular Cell Biology | Caltech | NIH Director’s Blog
Simultaneous generation of large numbers of strand-specific RNA-Seq cDNA libraries in a single reaction
Nature Methods
Shishkin AA, Giannoukos G, Kucukural A, Ciulla D, Busby M, Surka CF, Chen J, Bhattacharyya RP, Rudy RF, Patel MM, Novod N, Hung DT, Gnirke A, Garber M, Guttman M*, Jonathan Livny (2015).
Featured in: Nature Methods
RNA Antisense Purification (RAP) for mapping RNA interactions with chromatin
Methods in Molecular Biology
Engreitz JM, Lander ES, and Guttman M (2015).
The NIH BD2K center for big data in translational genomics
J Am Med Inform Assoc.
Paten B, Diekhans M, Druker BJ, Friend S, Guinney J, Gassner N, Guttman M, Kent WJ, Mantey P, Margolin AA, Massie M, Novak AM, Nothaft F, Pachter L, Patterson D, Smuga-Otto M, Stuart JM, Van’t Veer L, Wold B, Haussler D (2015).
2014
Long non-coding RNAs: An emerging link between gene regulation and nuclear organization
Trends in Cell Biology
Quinodoz S and Guttman M (2014).
RNA-RNA Interactions Enable Specific Targeting of Noncoding RNAs to Nascent Pre-mRNAs and Chromatin Sites
Cell
Engreitz JM, Sirokman K, McDonel P, Shishkin A, Surka CF, Russell P, Grossman SR, Chow AY, Guttman M, Lander ES (2014).
Transcriptome-wide Mapping Reveals Widespread Dynamic-Regulated Pseudouridylation of ncRNA and mRNA
Cell
Schwartz S, Bernstein DA, Mumbach MR, Jovanovic M, Herbst RH, León-Ricardo BX, Engreitz JM, Guttman M, Satija R, Lander ES, Fink G, Regev A (2014).
Methods for comprehensive experimental identification of RNA–protein interactions
Genome Biology
McHugh CA, Russell P, and Guttman M (2014).
Topological Organization of Multi-chromosomal Regions by Firre
Nature Structural & Molecular Biology
Hacisuleyman E, Goff LA, Trapnell C, Williams A, Henao-Mejia J, Sun L, McClanahan P, Hendrickson DG, Sauvageau M, Kelley DR, Morse M, Engreitz JM, Lander ES, Guttman M, Lodish HF, Flavell R, Raj A, and Rinn JL (2014).
2013
The Xist lncRNA Exploits Three-Dimensional Genome Architecture to Spread Across the X-chromosome
Science
Engreitz JM, Pandya-Jones A, McDonel P, Shishkin A, Sirokman K, Surka C, Kadri S, Xing J, Goren A, Lander ES, Plath K, and Guttman M. (2013).
Featured in: Science | Current Biology | Science News | Caltech
Ribosome Profiling Provides Evidence that Large Noncoding RNAs Do Not Encode Proteins
Cell
Guttman M, Russell P, Ingolia NT, Weissman JS, Lander ES. (2013).
Featured in: Nature Reviews Genetics
Transcriptional and Epigenetic Dynamics during Specification of Human Embryonic Stem Cells
Cell
Gifford CA, Ziller MJ, Gu H, Trapnell C, Donaghey J, Tsankov A, Shalek A, Kelley DR, Shishkin AA, Issner R, Zhang X, Coyne M, Fostel JL, Holmes L, Meldrim J, Guttman M, Epstein C, Park H, Kohlbacher O, Rinn JL, Gnirke A, Lander ES, Bernestin BE, Meissner A. (2013).
Mutations causing medullary cystic kidney disease type 1 lie in a large VNTR in MUC1 missed by massively parallel sequencing
Nature Genetics
Kirby A, Gnirke A, Jaffe DB, Baresova V, Pochet N, Blumenstiel B, Ye C, Aird D, Stevens C, Robinson JT, Cabili MN, Gat-Viks I, Kelliher E, Daza R, DeFelice M, Hulkova H, Sovova J, Vyletal P, Antignac C, Guttman M, Handsaker RE, Perrin D, Steelman S, Sigurdsson S, Scheinman SJ, Sougnez C, Cibulskis K, Parkin M, Green T, Rossin E, Zody MC, Xavier RJ, Pollack MR, Alper SL, Lindblad-Toh K, Gabriel S, Hart PS, Regev A, Nusbaum C, Kmoch S, Bleyer AJ, Lander ES, Daly MJ. (2013).
2012
A High-Throughput Chromatin Immunoprecipitation Approach Reveals Principles of Dynamic Gene Regulation in Mammals
Molecular Cell
Garber M, Yosef N, Goren A, Raychowdhury R, Thielke A, Guttman M, Robinson J, Minie B, Chevrier N, Itzhaki Z, Blecher-Gonen R, Bornstein C, Amann-Zalcenstein D, Weiner A, Friedrich D, Meldrim J, Ram O, Cheng C, Gnirke A, Fisher S, Friedman N, Wong B, Bernstein BE, Nusbaum C, Hacohen N, Regev A, Amit I. (2012)
2011
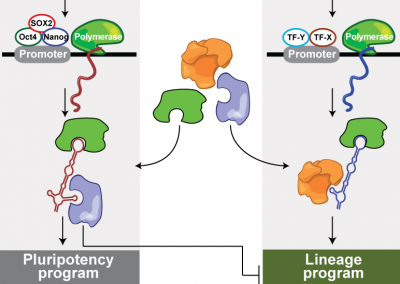
lincRNAs Act in the Circuitry Controlling Pluripotency and Differentiation
Nature
Guttman M, Donaghey J, Carey BW, Garber M, Grenier J, Munson G, Young G, Lucas AB, Ach R, Bruhn L, Yang X, Amit I, Meissner A, Regev A, Rinn JL, Root DE, Lander ES. (2011).
Featured in: Circulation Research | Cell Research
Evolutionary Constraint in the Human Genome Based on 29 Eutherian Mammals
Nature
Lindblad-Toh K, Garber M, Or Zuk, Lin MF, Parker BJ, Washietl S, Kheradpour P, Ernst J, Jordan G, Mauceli E, Ward LD, Lowe CB, Holloway AK, Clamp M, Gnerre S, Alfoldi J, Beal K, Chang J, Clawson H, Cuff J, Di Palma F, Fitzgerald S, Flicek P, Guttman M, Hubisz MJ, Jaffe DB, Jungreis I, Kent WJ, Kostka D, Lara M, Martins AL, Massingham T, Moltke I, Raney NJ, Rasmussen MD, Robinson J, Stark A, Vilella AJ, Wen J, Xie X, Zody MC, Worley KC, Kovar CL, Muzny DM, Gibbs RA, Warren WC, Mardis ER, Weinstock GM, Wilson RK, Birney E, Margulies EH, Herrero J, Green ED, Haussler D, Siepel A, Goldman N, Pollard KS, Pedersen JS, Lander ES, Kellis M. (2011).
Computational Methods for Transcriptome Annotation and Quantification using RNA-Seq
Nature Methods
Garber M, Grabherr MG, Guttman M, Trapnell C. (2011).
2010
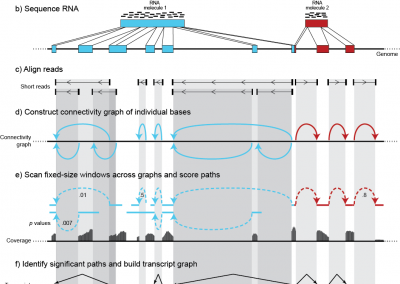
Ab initio Reconstruction of Transcriptomes of Pluripotent and Lineage Committed Cells Reveals Gene Structures of Thousands of lincRNAs
Nature Biotechnology
Guttman M, Garber M, Levin JZ, Donaghey J, Robinson J, Adiconis X, Fan L, Koziol MJ, Gnirke A, Nusbaum C, Rinn JL, Lander ES, Regev A. (2010)
Featured in: Nature Biotechnology
A Large intergenic Noncoding RNA Induced by p53 Mediates Global Gene Repression in the p53 Transcriptional Response
Cell
Huarte M, Guttman M, Feldser D, Garber M, Koziol M, Broz D, Khalil AM, Zuk O, Amit I, Rabani M, Attardi L, Regev A, Lander ES, Jacks T, Rinn JL. (2010)
Integrative Genomics Viewer
Nature Biotechnology
Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. (2010).
Large Intergenic Non-coding RNA-RoR Modulates Reprogramming of Human Induced Pluripotent Stem Cells
Nature Genetics
Loewer S, Cabili MN, Guttman M, Loh YH, Thomas K, Park IH, Garber M, Curran M, Onder T, Agarwal S, Manos PD, Datta S, Lander ES, Schlaeger TM, Daley GQ, Rinn JL. (2010).
Digital Transcriptome Profiling from Attomole-level RNA Samples
Genome Research
Ozsolak F, Goren A, Gymrek M, Guttman M, Regev A, Bernstein BE, Milos PM. (2010)
2009
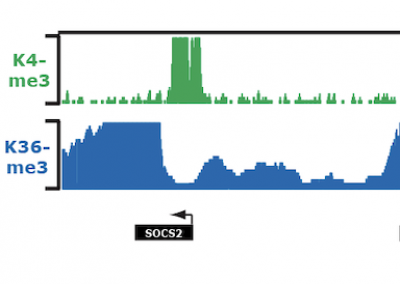
Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals
Nature
Guttman M, Amit I, Garber M, French C, Lin M, Feldser D, Huarte M, Cabili M, Carey BW, Cassady J, Jaenisch R, Mikkelsen T, Jacks T, Hacohen N, Bernstein BEB, Kellis M, Regev A, Rinn JL, Lander ES. (2009)
Featured in: Nature Biotechnology | Science
Many Human Large Intergenic Noncoding RNAs Associate with Chromatin-Modifying Complexes and Affect Gene Expression
Proceedings of the National Academies of Science
Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, Regev A, Lander ES, Rinn JL. (2009)
Unbiased Reconstruction of a Mammalian Transcriptional Network Mediating Pathogen Responses
Science
Amit I, Garber M, Chevrier N, Leite AP, Donner Y, Eisenhaure T, Guttman M, Grenier JK, Li W, Zuk O, Schubert LA, Birditt B, Shay T, Goren A, Zhang X, Smith Z, Deering R, McDonald RC, Cabili M, Bernstein BE, Rinn JL, Meissner A, Root DE, Hacohen N, Regev A. (2009)
Identifying Novel Constrained Elements by Exploiting Biased Substitution Patterns
Bioinformatics
Garber M, Guttman M, Clamp M, Zody MC, Friedman N, Xie X. (2009).

Ready to Collaborate?
Contact Us