Technologies
SPRITE
Split-Pool Recognition of Interactions by Tag Extension (SPRITE) is a method to map higher-order 3D molecular interactions in the nucleus. This method identifies interactions that occur across larger spatial distances than can be observed by proximity ligation-based methods and enables genome-wide detection of multiple RNA and DNA interactions that occur simultaneously.
Briefly, SPRITE works as follows: DNA, RNA, and protein are crosslinked in cells; nuclei are isolated; chromatin is fragmented; interacting molecules within an individual complex are barcoded using a split-pool strategy; and interactions are identified by sequencing and matching all reads that contain identical barcodes.
DNA SPRITE
DNA SPRITE enables genome-wide detection of multiple simultaneously occurring higher-order DNA interactions within the nucleus and provides a global picture of inter-chromosomal spatial organization – including around nuclear bodies.
RNA & DNA SPRITE
RNA & DNA SPRITE (RD-SPRITE) simultaneously measures RNA-RNA, RNA-DNA, and DNA-DNA contacts within 3D nuclear structures and allows for comprehensive mapping of the location of all classes of RNA (including non-coding, mRNAs, and nascent RNA) relative to DNA and other RNA molecules.
Single Cell SPRITE
Single Cell SPRITE (scSPRITE) measures higher-order nucleic acid interactions in thousands of individual nuclei simultaneously and enables generation of high-resolution genome structure maps within individual cells.

ChIP-DIP
ChIP-DIP (ChIP Done In Parallel) is a split-pool based method that enables simultaneous, genome-wide mapping of hundreds of diverse regulatory proteins to genomic DNA within a single experiment.


CLAP
Covalent Linkage & Affinity Purification (CLAP) is a method to stringently identify and precisely map RNA-protein interactions in vivo. CLAP integrates an epitope tag into a protein of interest which enables covalent coupling of the protein to a resin (e.g. HaloTag, SpyTag) thereby enabling a purification with fully denaturing conditions – including high temperatures, high concentrations of denaturants and detergents, and chaotropic salts – that disrupt protein and RNA folding. CLAP eliminates RNA-protein interactions that occur in solution and retains in vivo RNA-protein interactions detected by methods such as CLIP.


RAP
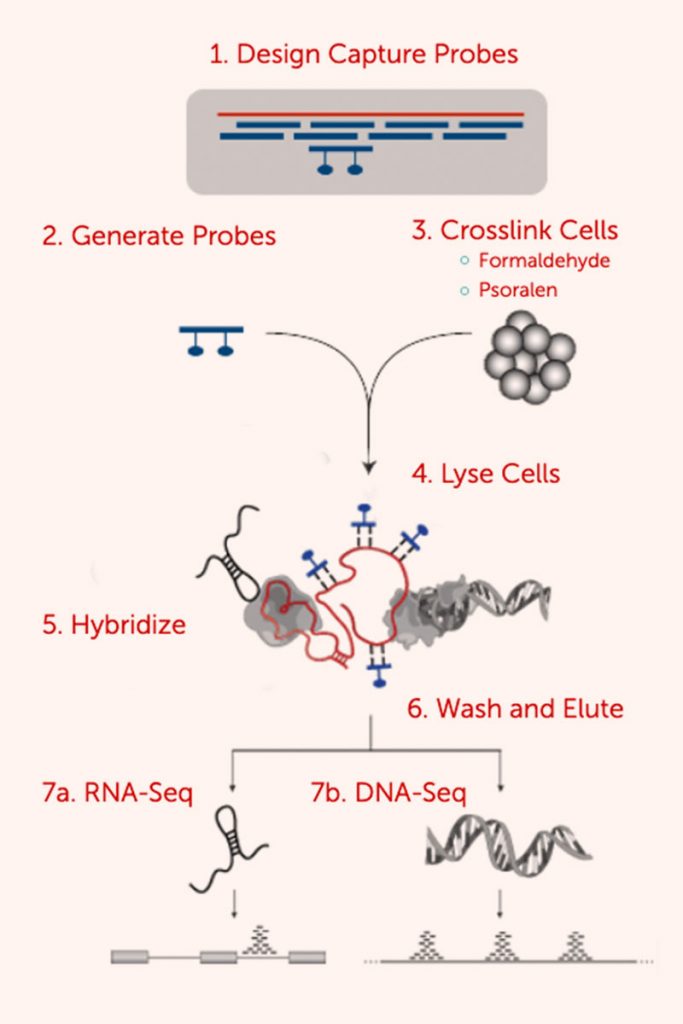
RNA Antisense Purification (RAP) is a method to purify long non-coding RNA (lncRNA) complexes in vivo. RAP uses biotinylated antisense probes to hybridize to a target RNA to purify the endogenous RNA and its associated proteins, RNA, and genomic DNA from crosslinked cell lysate. We designed RAP to enable specific purification of chromatin associated with a target lncRNA, achieve high resolution mapping of the associated DNA target sites upon sequencing of the captured DNA, and robustly capture any lncRNA with minimal optimization.
RAP was developed by Jesse Engreitz with help from Patrick McDonel, Alex Shishkin, Klara Sirokman, and Christine Surka.
RAP-MS
RNA Antisense Purification with Mass Spectrometry (RAP-MS) is a method to purify a lncRNA complex in vivo and identify the direct interacting proteins by quantitative mass spectrometry. RAP-MS uses UV crosslinking to create covalent linkages between directly interacting RNA and protein and purifies lncRNAs in denaturing conditions to disrupt non-covalent interactions.

RNAtag-Seq
RNAtag-Seq is a method for generating a single RNA-Seq library from a large number of independently barcoded RNA samples. This method provides highly reproducible, strand-specific, quantitative sequencing covering the full lengths of transcripts in diverse species. This approach dramatically increases the throughput and decreases the cost per sample of RNA-Seq library construction.


